



Leveraging on the OHDSI/ OMOP CDM to develop a systematic approach to clinical research for answering real-world questions across a global network of standardized Covid-19 data in Kenya and Malawi.
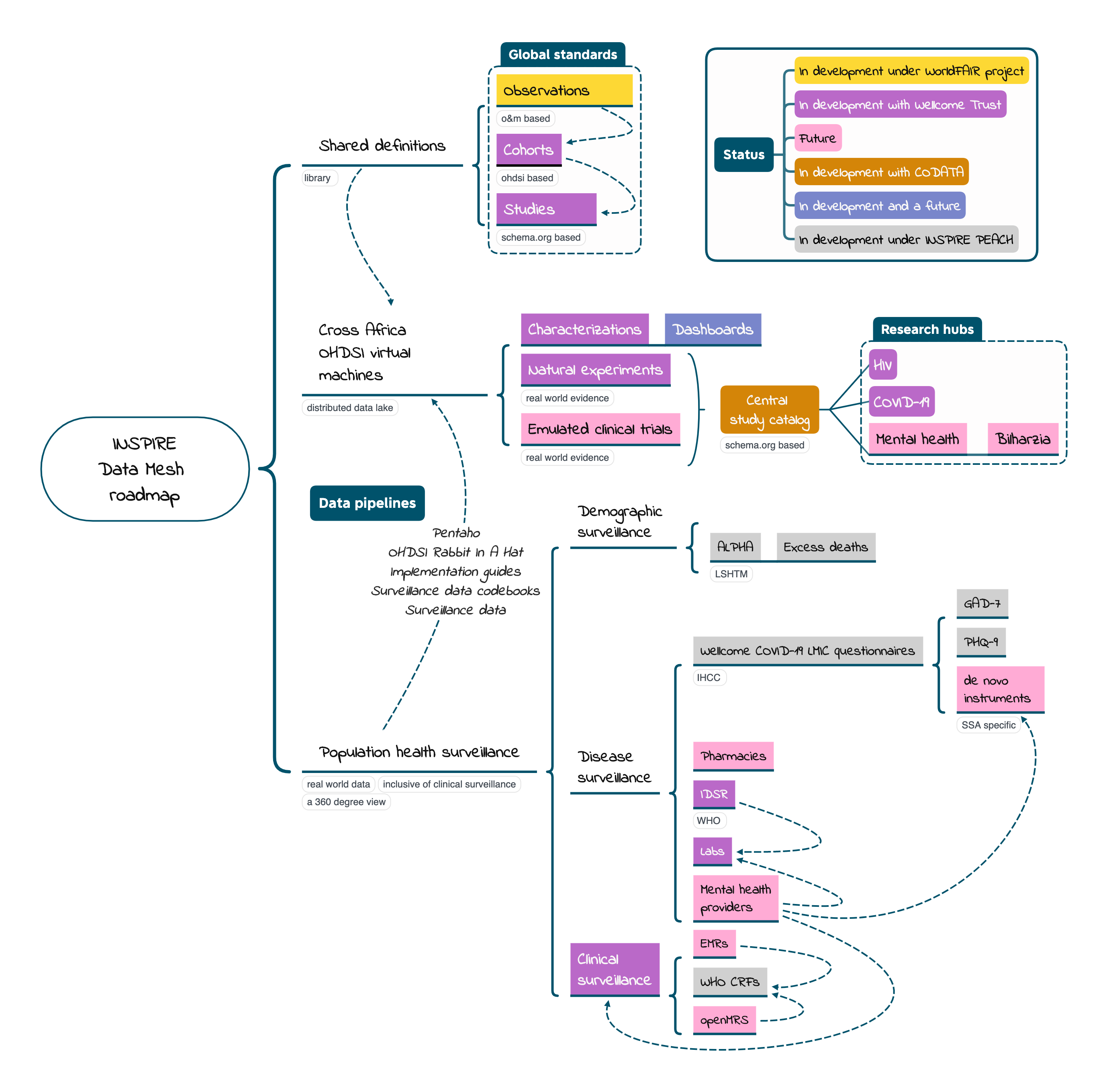
In a data mesh, it is the shared definitions across a federation that assures that local research can be combined across space and time as needed. In the context of OHDSI and OMOP there are shared definitions on several levels.
Cross-Africa virtual machines consume population health surveillance data in line with the shared definitions first to characterize the data with the assistance of dashboards and then to execute the research.
OHDSI virtual machines (VMs) are constructed under the same or different cloud providers and contain the OMOP CDM and a set of OHDSI services that run on top of OMOP. Currently INSPIRE has built its own VM that runs at just one cloud provider – Microsoft Azure. Both INSPIRE and other OHDSI research entities are in the process of building VMs that, through containerization, can run under multiple cloud providers.
INSPIRE imagines that over time a brigade of VMs will be deployed in the African context that are birthed by one or more research entities. Membership in the brigade will be coordinated by the Africa Chapter of OHDSI. Each VM will have to pass the same test.
INSPIRE is building a catalog that chronicles OHDSI experiments undertaken across VMs. The catalog is a collection of observational studies specified with schema.org. Each study published at the INSPIRE website will appear in an internet-wide catalog of datasets called Google Dataset Search. Google Dataset Search is, in effect, a catalog of catalogs.
Data pipelines facilitate the movement of (meta)data from Population Health Surveillance (below) into an OMOP CDM instance (VM). There are specific pipelines for each type of surveillance – demographic, disease, clinical (including COVID-19) and, in the future, sensors that capture environmental exposures.
Data pipelines are centrally managed shared resources across OMOP CDM instances.
While these pipelines differ in detail depending on the surveillance type, at a high level they all follow the same steps:
The VMs consume population health surveillance marshaled into multiple exchange formats aka standards. These exchange standards and the data they format are run through shared data pipelines at each VM.
At INSPIRE population health surveillance is growing incrementally. This means we have a roadmap, and we are adding new data pipelines in connection with new data types over time. Currently, INSPIRE supports the following data types, more or less: